BLOG > Publications & Citations > TRACERx analysis identifies a role for FAT1 in regulating chromosomal
Authors: Lu, WT., Zalmas, LP., Bailey, C. et al.
Source: Nat Cell Biol 27, 154–168 (2025).
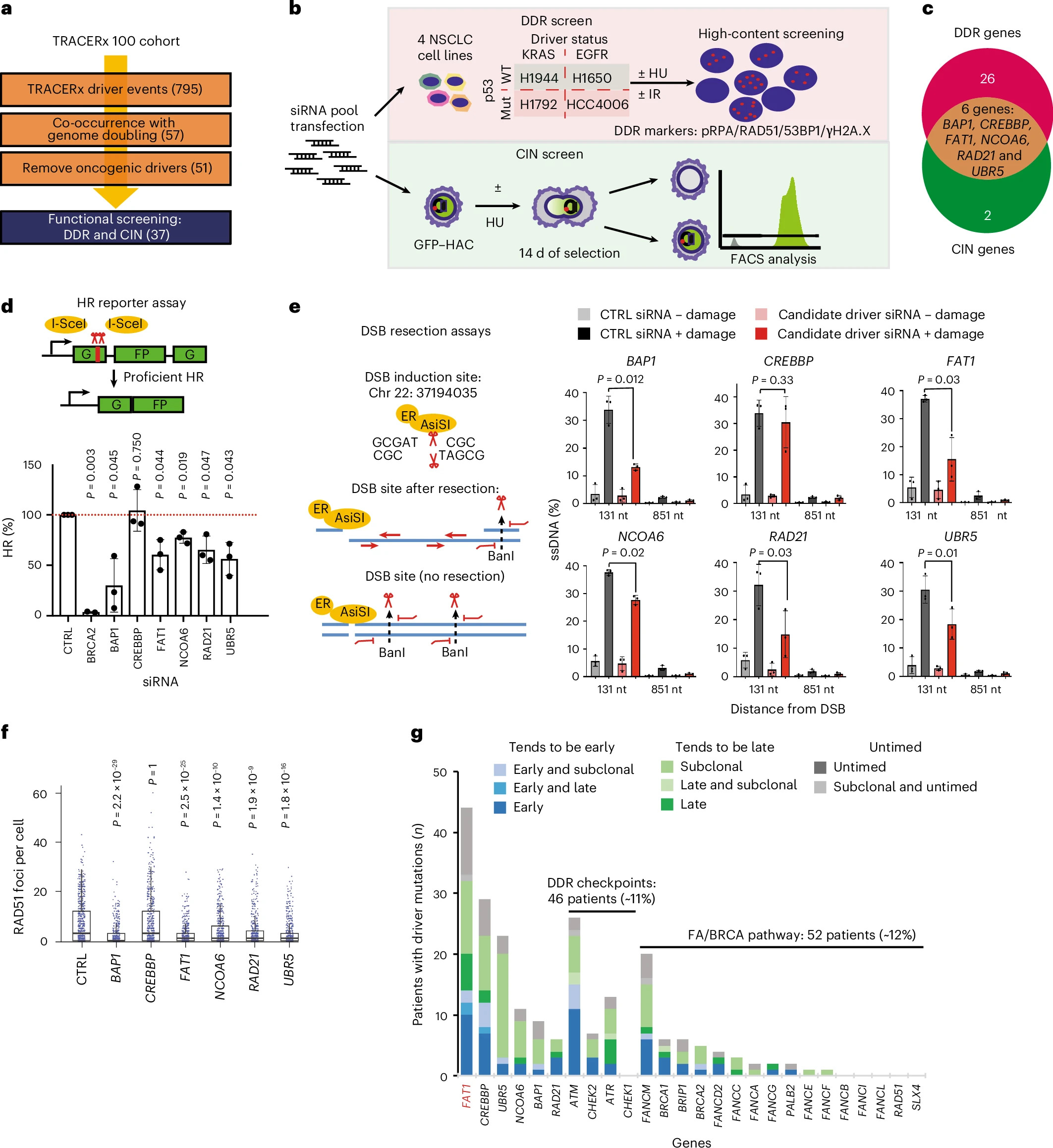
We are thrilled to share insights from a recent study entitled "TRACERx analysis identifies a role for FAT1 in regulating chromosomal instability and whole-genome doubling via Hippo signalling" published in Nature Cell Biology by Wei-Ting Lu, Lykourgos-Panagiotis Zalmas, Chris Bailey et al.
"Chromosomal instability (CIN) is common in solid tumours and fuels evolutionary adaptation and poor prognosis by increasing intratumour heterogeneity. Systematic characterization of driver events in the TRACERx non-small-cell lung cancer (NSCLC) cohort identified that genetic alterations in six genes, including FAT1, result in homologous recombination (HR) repair deficiencies and CIN. Using orthogonal genetic and experimental approaches, we demonstrate that FAT1 alterations are positively selected before genome doubling and associated with HR deficiency. FAT1 ablation causes persistent replication stress, an elevated mitotic failure rate, nuclear deformation and elevated structural CIN, including chromosome translocations and radial chromosomes. FAT1 loss contributes to whole-genome doubling (a form of numerical CIN) through the dysregulation of YAP1. Co-depletion of YAP1 partially rescues numerical CIN caused by FAT1 loss but does not relieve HR deficiencies, nor structural CIN. Importantly, overexpression of constitutively active YAP15SA is sufficient to induce numerical CIN. Taken together, we show that FAT1 loss in NSCLC attenuates HR and exacerbates CIN through two distinct downstream mechanisms, leading to increased tumour heterogeneity."
Congratulations to all authors for this great article.
Our Lullaby Stem was used to perform gene silencing in H1994 cells with siRNA.
Read the article See our Lullaby Stem