BLOG > mRNA & LNP > Messenger RNA: from transcription to translation
Summary:
1. Understanding mRNA: definition & structure
2. Transcription: the initiation of mRNA synthesis
3. mRNA splicing & export, nuclear transport and regulation
4. mRNA translation: decoding codons into proteins
1. Understanding mRNA: definition & structure
mRNA definition
Messenger RNA (mRNA), is a single-stranded RNA molecule transcribed from a DNA template during gene expression. It functions as an intermediate, conveying the genetic instructions encoded in DNA to the ribosomes, where protein synthesis occurs.
Cellular function of mRNA
The primary function of mRNA is to act as a template for protein biosynthesis. Following transcription in the nucleus, mRNA transports the genetic code to the cytoplasm, where ribosomes decode the nucleotide sequence into a specific amino acid chain. This translation process ensures the accurate production of proteins according to the genetic blueprint. Beyond its role in translation, mRNA also contributes to the regulation of gene expression through mechanisms such as mRNA stability, localization, and interaction with microRNAs or RNA-binding proteins.
mRNA structure
mRNA is composed of a linear sequence of ribonucleotides, each containing a ribose sugar, a phosphate group, and a nitrogenous base. Just like DNA, but with one key difference: mRNA incorporates uracil (U) instead of thymine (T). This single-stranded structure makes mRNA highly versatile and readily accessible for ribosomal translation.
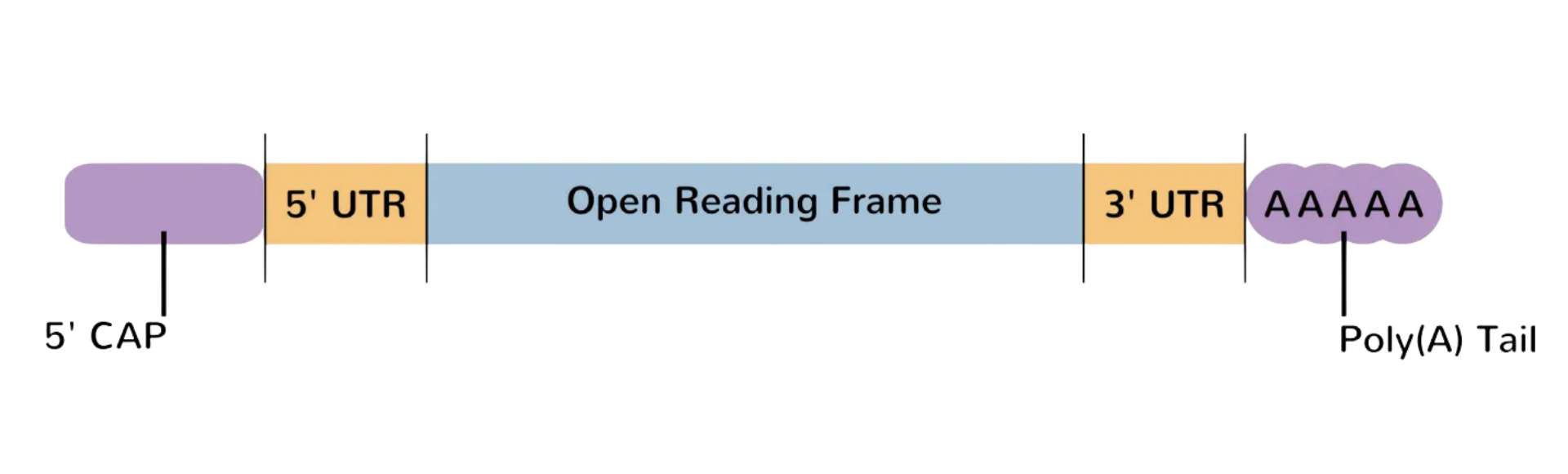
5’ Cap
The 5' cap is a modified guanine nucleotide (m⁷G) attached via a 5' to 5' triphosphate linkage to the 5' end of eukaryotic mRNA, forming a protective structure. This cap includes methylation at the 7th position of guanine (cap-0), with additional 2'-O-methylations on the first ribose (cap-1) or first two riboses (cap-2) in higher eukaryotes, enhancing stability and immune recognition. It prevents degradation by 5' exonucleases by mimicking a 3' end and recruits cap-binding proteins like the CBC (cap-binding complex) to regulate nuclear export. The cap also promotes translation initiation by interacting with eIF4E/eIF4G factors and facilitates 5' proximal intron excision during splicing.
In in vitro workflows, enzymatic capping can be performed using our Capping Kit. This kit permits the addition of a Cap0 or Cap1 structure on the 5' terminus of mRNA, improving its stability and translation.
Order Capping Kit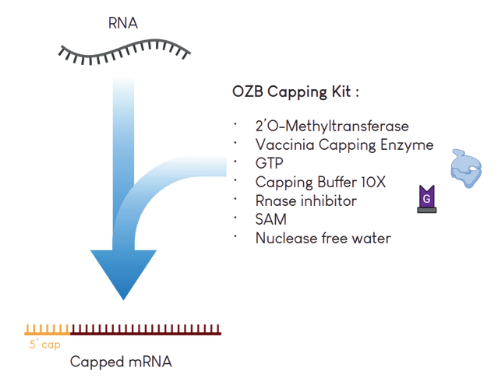
5’ Untranslated Region (5’ UTR)
The 5’ untranslated region (5’ UTR) is a non-coding region directly upstream from the initiation codon involved in the post- transcriptional regulation of gene expression by modulating mRNA stability, transport, subcellular localization and translation efficiency thus allowing a fine control of the protein product. This region has a high GC content and several secondary structures and comprises the Kozak sequence (GCCGCCRCCAUGG) that plays a major role in the initiation of the translation process. The structure and sequence of the 5’ UTR, including the Kozak sequence, critically influence ribosome binding and protein expression.
Open Reading Frame (ORF)
The open reading frame (ORF) of mRNA is the protein-coding region extending from the start codon (AUG) to an in-frame stop codon (UAA, UAG, or UGA). It specifies the amino acid sequence during translation and excludes untranslated regions (UTRs) flanking the coding sequence. In eukaryotes, the primary ORF is typically the longest sequence lacking stop codons, while prokaryotic mRNAs may contain multiple ORFs. ORF identification aids gene prediction, as conserved, lengthy sequences often indicate functional protein-coding regions.
3’ Untranslated Region (3’UTR)
The 3’ untranslated region (3’ UTR) of mRNA is the segment located downstream of the stop codon, extending to the poly(A) tail. It does not encode protein but contains regulatory elements that influence mRNA stability, localization, export and translation efficiency. The 3’ UTR often serves as a binding site for microRNAs and RNA-binding proteins, modulating post-transcriptional gene expression.
Poly(A) tail
The poly(A) tail is a sequence of adenosine residues, typically 50 to 250 nucleotides long with a median length of around 100–150 nucleotides, added to the 3’ end of eukaryotic mRNA after transcription. The poly(A) tail contains binding sites for poly(A) binding proteins (PABPs) that play a major role in export from the nucleus, translation, and protection from degradation. Its length is an important determinant of translational efficiency and mRNA stability. This is an important element as its absence or removal often leads to exonuclease-mediated degradation of the mRNA.
The resulting polyadenylated RNA is more stable and translation-ready, ideal for downstream use in research or therapeutic models.
Order Poly(A) Tailing Kit-kit.png)
2. Transcription: the initiation of mRNA synthesis
Process of transcription
Transcription is the biological process by which a segment of DNA is transcribed into a complementary messenger RNA (mRNA) strand by the enzyme RNA polymerase. The process begins with initiation, where RNA polymerase binds to a promoter region on the DNA and unwinds the double helix to expose the template strand. During the elongation phase, RNA polymerase reads the DNA template strand and assembles a complementary RNA sequence. In this process, adenine (A) pairs with uracil (U), and cytosine (C) pairs with guanine (G). In eukaryotes, this primary RNA transcript undergoes processing, including 5’ capping, splicing, and addition of a poly(A) tail, to become mature mRNA. The mature mRNA then exits the nucleus and serves as a template for protein synthesis in the cytoplasm. Transcription is tightly regulated, ensuring that genes are expressed at the right time and in appropriate amounts.
In vitro transcription (IVT)
RNA can be synthesized in vitro by the so-called in vitro transcription (IVT) technique (figure 2). Linearized plasmid DNA, PCR products or cDNA can be used as templates for transcription if they contain a double-stranded RNA polymerase promoter region in the correct orientation with consensus promoter sequences of different RNA Polymerases T7, T3 or SP6. RNA will be synthesized by action of a T7 polymerase. The common size of IVT mRNAs ranges between 500 to 5000 nucleotides, although smaller or larger RNAs can be synthesized upon protocol adaptation.
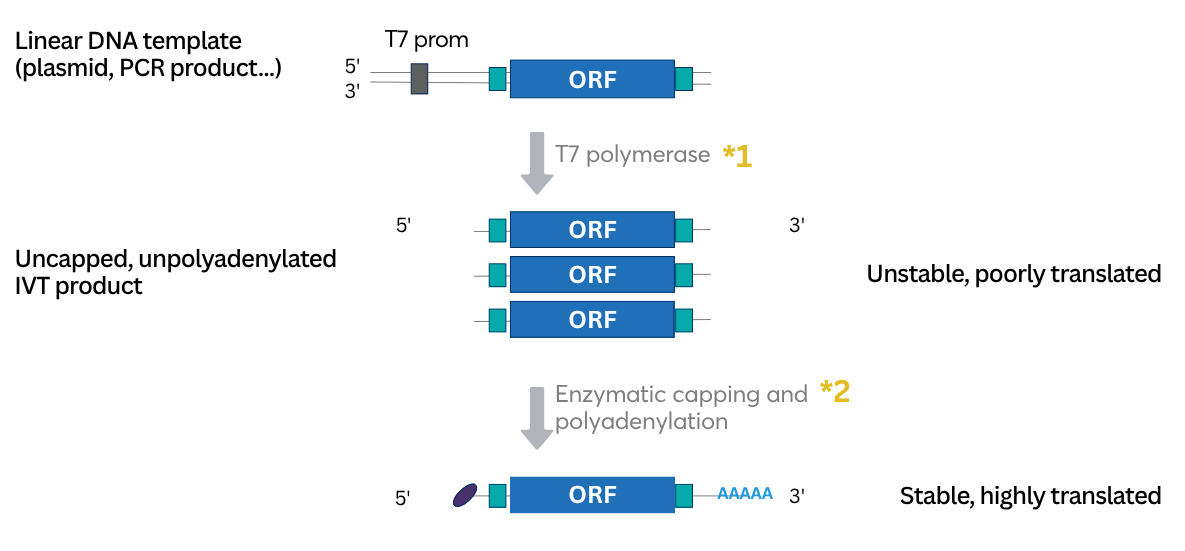
Figure 2: Scheme of synthetic RNA production by in vitro transcription.
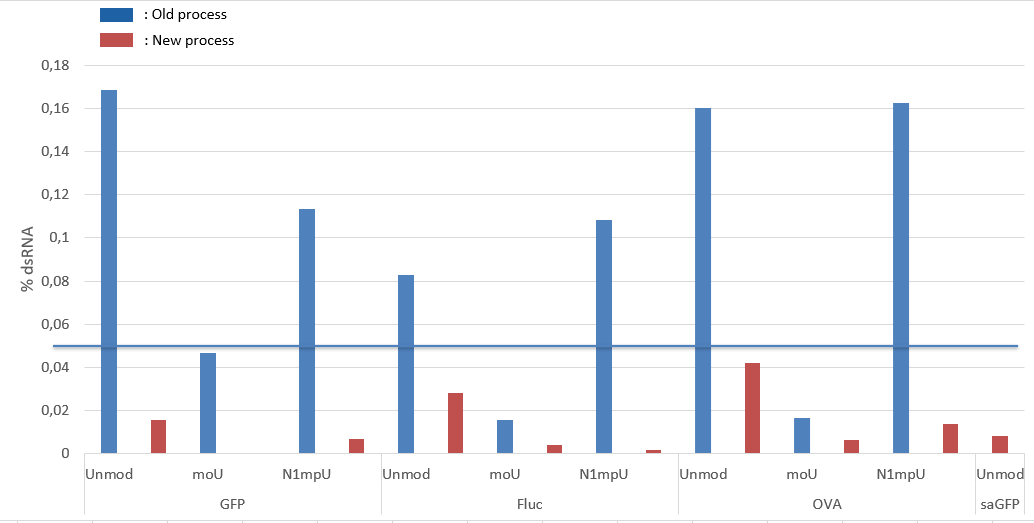
To evade innate immune detection, modified nucleoside triphosphates (NTPs), such as pseudouridine, thiouridine, N1-methylpseudouridine, or 5-methoxyuridine (moU), can be substituted for the uridine triphosphate during IVT. Indeed, replacement of only 25% of uridine and cytidine with 2-thiouridine and 5-methyl-cytidine synergistically decreased mRNA binding to pattern recognition receptors, such as TLR3, TLR7, TLR8 and RIG-I, in human peripheral blood mononuclear cells (PBMCs).
In addition, double stranded RNA (dsRNA) impurities formed during transcription significantly contribute to the innate immune activity of mRNA therapeutics. Purification of IVT mRNA by means of high-performance liquid chromatography (HPLC), or alternative simplified methods using cellulose columns or selective digestion of dsRNA fragments using RNAse III have been established and proven to greatly reduce immune reaction.
To support such workflows, we provide an IVT Kit optimized for high-yield mRNA synthesis. Designed to transcribe linearized plasmid DNA or PCR fragments, this kit yields approximately 100 µg of RNA per µg of DNA template, depending on sequence length and purity.
The RNA generated using this kit can be employed in a diverse range of experiments such as in vitro translation, cell transfection, isolation of RNA binding proteins, etc...
Order IVT Kit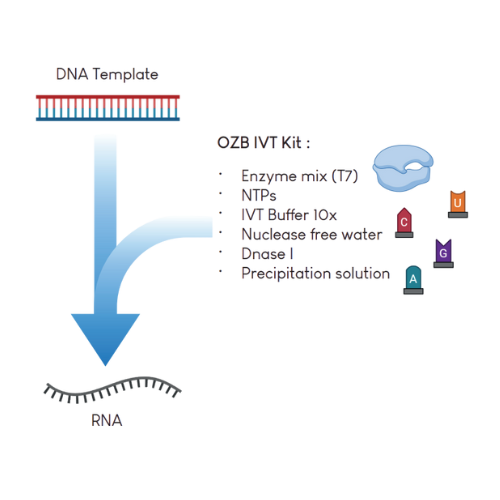
3. mRNA splicing & export, nuclear transport and regulation
mRNA Splicing
Splicing of pre-mRNA typically occurs in the nucleus either during (co-transcriptionally) or immediately after transcription. It removes non-coding introns and joins exons to produce a continuous coding sequence. This process is critical in eukaryotic gene expression and can also generate alternative transcript isoforms through regulated splicing patterns. While splicing is not typically a concern in synthetic mRNA workflows (as IVT templates are usually intron-free), understanding this mechanism is key to designing coding sequences that reflect native transcript structures.
mRNA Export
After processing in the nucleus, mature mRNA assembles into messenger ribonucleoprotein complexes (mRNPs) and is recognized by export factors. These complexes are actively transported through the nuclear pore complex (NPC) via interactions between export receptors and FG-nucleoporins. Upon reaching the cytoplasmic side, remodeling factors such as Dbp5 and Gle1 dissociate export components, releasing the mRNA into the cytoplasm for translation. Once translocated through the NPC into the cytoplasm, mRNA is released from the export machinery and can be directed toward specific subcellular regions. This localization relies on cis-acting RNA localization elements, typically located in the 3' untranslated region (3' UTR), which are recognized by RNA-binding proteins. These elements contribute not only to the spatial regulation of translation, but also to mRNA stability and degradation.
The role of the nuclear pore complex (NPC)
Export is facilitated by the nuclear pore complex (NPC), a massive multiprotein assembly embedded in the nuclear envelope. The NPC serves as a selective gateway, controlling the bidirectional transport of macromolecules between the nucleus and cytoplasm. Due to their size and structural complexity, mRNA molecules cannot passively diffuse through the NPC and therefore require the assistance of export receptors.
4. mRNA translation: decoding codons into proteins
Ribosome function and codon recognition
Translation is carried out by ribosomes, which decode the mRNA sequence in the 5′ to 3′ direction to synthesize protein. It begins with the assembly of the ribosome at the start codon of the mRNA and the recruitment of the initiator tRNA. Each codon (comprising three-nucleotide sequence) corresponds to a specific amino acid, defined by the universal genetic code. There are 64 possible codons, each encoding for a specific amino acid or signaling the end of protein synthesis. The ribosome ensures correct pairing between mRNA codons and tRNA anticodons, driving polypeptide elongation.
Stages of Translation
- Initiation: begins with ribosome assembly at the start codon (typically AUG), guided by initiation factors and the 5′ cap structure.
- Elongation: proceeds as the ribosome translocates along the mRNA, catalyzing peptide bond formation between successive amino acids delivered by charged tRNAs.
- Termination: occurs when a stop codon (UAA, UAG, or UGA) is encountered; release factors promote polypeptide release and ribosome disassembly.
While at this stage of RNA preparation, the molecules are fully mature, rather stable and translatable, mRNAs are still immunogenic. Altering the mRNA’s base composition is an established method that consists in reducing immunogenicity for mRNA therapeutics. Since uridine-rich sequences of RNA trigger the innate immune response, depleting uridine triphosphate from the final mRNA sequence can improve evasion. Substituting a U-containing codon during sequence optimization for one without U will result in the same downstream protein while reducing the mRNA immunogenicity.
To learn more about mRNA, visit our other ressources:
Production kits mrnaCatalog mrna